About Prof. R. (Roland) Kanaar
Introduction
Academic career
Roland Kanaar studied chemistry at Leiden University and obtained his PhD degree in 1988 for research on the action of an enhancer in site-specific DNA recombination and the elucidation of how nucleoprotein complexes assembled at distant sites along a DNA chain communicate with each other to provide selectivity during recombination. His post-doctoral work with at the University of California, Berkeley, aimed at understanding mechanisms of homologous recombination (with Dr. Nick R. Cozzarelli) and at understanding how proteins and RNA interact to achieve accurate but flexible recognition of splice sites (with Dr. Don Rio). In 1995 he joined the Department of Genetics and the Department of Radiation Oncology at Erasmus University Medical Center Rotterdam. In 2000 he was appointed Professor of Molecular Radiation Genetics. His current research addresses the mechanisms and biological relevance of the DNA damage response. The DNA damage response is essential to prevent chromosomal abnormalities, which in their turn may lead to hereditary diseases, cancer, cell decay and aging.
Interdisciplinary approach
A salient feature of his work is the strong multi- and interdisciplinary approach, which contributed to making him a leader in the field. His work spans the experimental range from single molecule biophysics to animal models. Depending on the specific question at hand techniques from biophysics, structural biology, biochemistry, cell biology, molecular biology and animal genetics are applied in his laboratory. Integrating information through this spectrum of techniques has a synergistic effect on the ability of his research group to translate fundamental insight into how the DNA damage response operates to how it can be used to select patients for DNA damage response-targeting precision cancer therapy and how it can be manipulated to design and explore novel cancer treatments.
European Molecular Biology Organization
In 2002 he was elected as a member of the European Molecular Biology Organization and in 2013 we was elected to the the Royal Netherlands Academy of Arts and Sciences (KNAW).
He has coordinated a number of large EU projects, including ‘DNA damage response and breast cancer’. He is a co-founder of Cancer Genomics Netherlands, which was established in 2013 as a ‘Center of Excellence’ through the Gravitation (Zwaartekracht) Program.
 |
 |
 |
Field(s) of expertise
Research
The DNA damage response: From molecular mechanism to application in cancer therapy
Our research team pursues two integrated lines of research:
(1) Mechanisms of the DNA damage response
(2) The impact and application of the DNA damage response in cancer
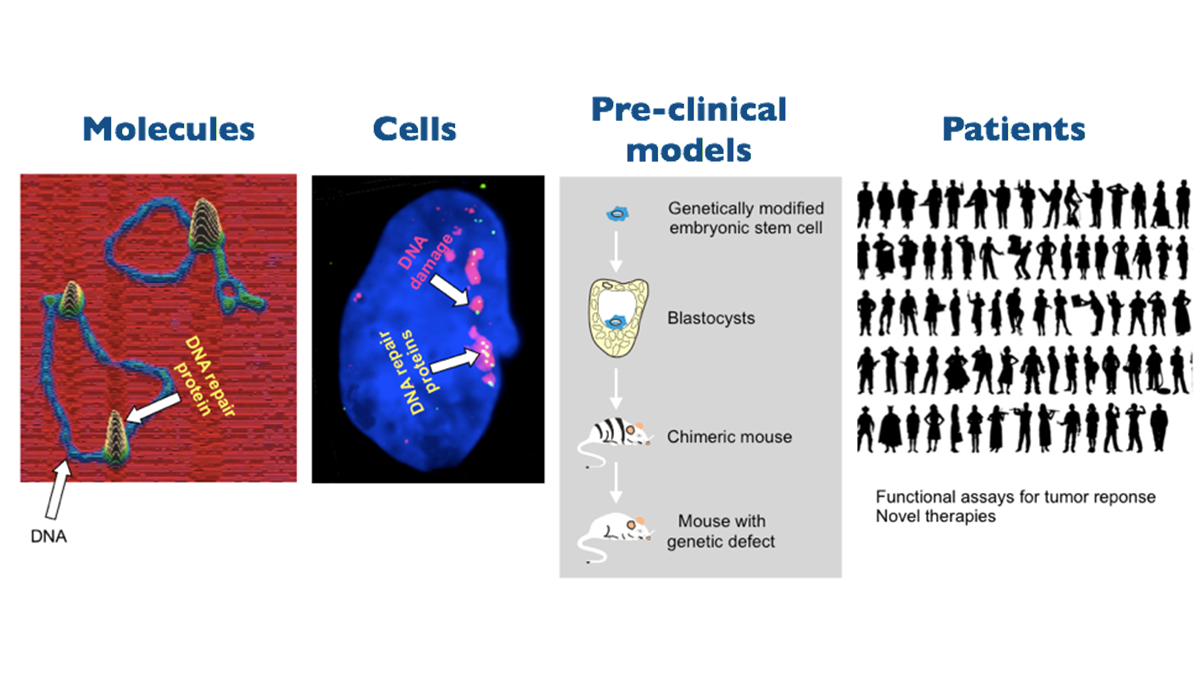
(1) Mechanisms of the DNA damage response
This research line addresses the fundamental mechanistic aspects of the DNA damage response with special attention to DNA repair by homologous recombination. We take complementary and interwoven approaches at both the molecular and cellular level. The molecular mechanisms of the multiple pathways of the DNA damage response are studied by applying and developing (single molecule) biochemical analyses, advanced proteomic analyses and state-of-the-art molecular imaging techniques to identify dynamic molecular interactions that are responsible for the assembly and disassembly of the molecular factories that guard and repair the genome. At the cellular level the DNA damage repair processes in (live) cells are monitored in real time for how they detect DNA damage and proceed to assemble and disassemble the repair machinery by applying the latest super-resolution techniques to understand the interplay between different DNA damage response pathways and their integration in the physiology of the cell.
The ultimate goal is be to define and dissect the molecular circuits of DNA damage repair and their integration in cellular processes. This knowledge is pivotal for identifying molecular targets for designing novel mechanism-based interventions and manipulations of these processes. This will provide the basis for improving existing approaches and developing entirely novel and innovative precision therapies for cancer and aging-related diseases.
(2) The impact and application of the DNA damage response in cancer
Many important anti-cancer treatments are based on the cell killing properties of DNA-damaging agents. The efficacy of such treatments depends on characteristics of the DNA damage response in tumor cells, which is almost invariably compromised and can be exploited to develop better patient selection methods, to guide targeted cancer treatments and to design novel precision cancer treatments.
Functional imaging of biological processes in pre-clinical cancer models
Custom-made genetically engineered pre-clinical mouse models and orthotopic tumor mouse models are developed to test treatments that are rationally designed based on molecular mechanistic insights from the ‘Mechanisms of the DNA damage response’ research line. This involves unique Erasmus MC resources for 3D imaging to identify and locate molecular processes in action in vivo in live mice in a longitudinal manner. The ultimate goal, in combination with the ex vivo functional assays on patient-derived materials described below, should be to develop (biology-based) image-guided, adaptive and targeted cancer therapies.
The DNA damage response as a functional tool in precision cancer therapy
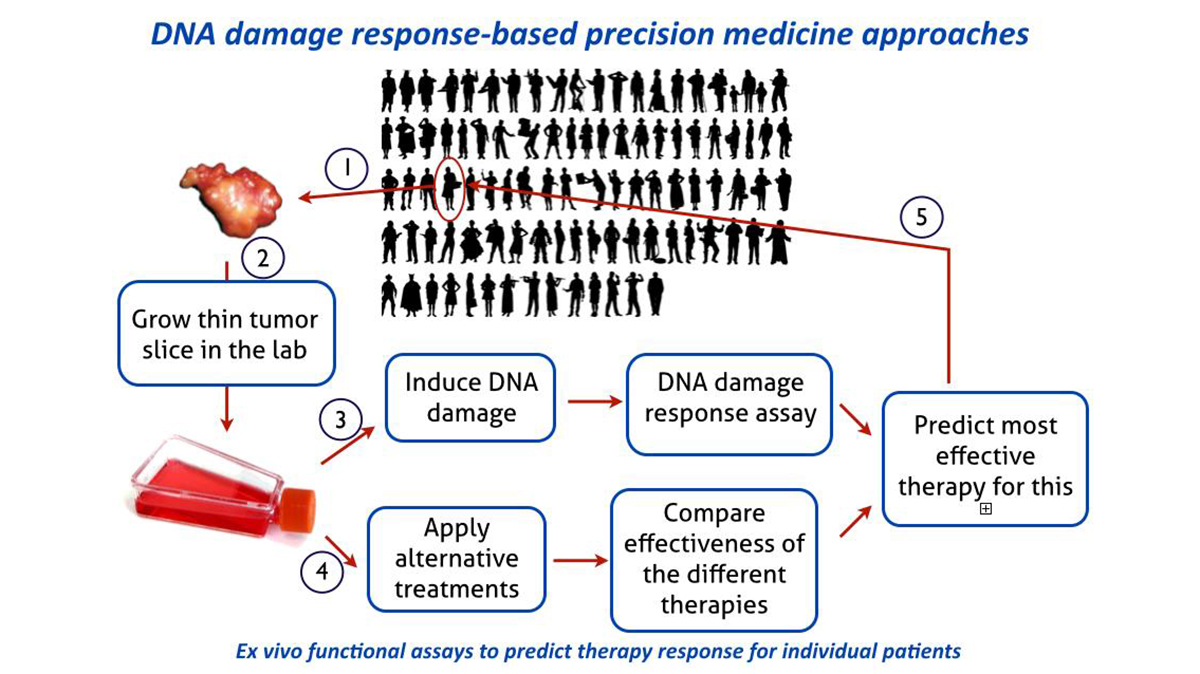
A major goal within this research line should be to obtain a direct functional appraisal of the DNA damage response capacity for individual patient tumors. Based on this principle, functional assays will be developed for multiple DNA damage response reactions and extend beyond breast cancer to other tumor sites. These assays will ultimately identify specific defects in DNA damage response that can be exploited in the exciting new strategy of synthetic lethality approaches, as pioneered for hereditary breast and ovarian cancer. The potential for synthetic lethality is in principle general and needs to be systematically explored in general for other tumors or for other DNA damage response defects and in the context of mechanism-based chronotherapeutic cancer treatment protocols. Connecting this research line to the Human Organ and Disease Model Technology (hDMT) Institute provides opportunities to adopt the functional DNA damage response assays on tumors to a microfluidic chip format for higher throughput greatly enhancing the utility in a clinical setting. By focusing research around precision therapies the Department will easily connect to the ‘Nationale Wetenschaps Agenda’ exemplar route ‘Personalized Medicine’.
Proton therapy
Furthermore, our team is initiating research on the DNA damage response towards protons through the close and long-standing collaboration with the Department of Radiation Oncology and with a collaboration to be initiated with Holland PTC, in the context of Medical Delta. Understanding the molecular mechanism and cellular pathways of the DNA damage response in reaction to protons will provide knowledge not only for enlarging the therapeutic window of proton therapy, but also for designing synergizing combination therapies.
Education and career
- 2015 - present
Director Joint Erasmus MC / TU Delft MSc program Nanobiology - 2000 - present
Professor of Molecular Radiation Genetics, Erasmus Medical Center, Dept. Molecular Genetics and Dept.Radiation Oncology, Rotterdam, The Netherlands - 1995 - 2000
Fellow of the Royal Netherlands Academy of Arts and Sciences, Erasmus University Rotterdam, Dept. Genetics, The Netherlands - 1992 - 1995
Senior postdoctoral fellow of The American Cancer Society, Dept. Molecular & Cell Biology, UC Berkeley, USA - 1990 - 1992
Postdoctoral fellow of The Jane Coffin Childs Memorial Fund for Medical Research, Dept. Molecular & Cell Biology, UC Berkeley, USA - 1989 - 1990
National Science Foundation postdoctoral fellow, Dept. Molecular Biology, UC Berkeley, USA - 1984 - 1988
Ph.D. Molecular Genetics, Leiden University, The Netherlands - 1982 - 1984
M.Sc. Biochemistry, Leiden University, The Netherlands - 1979 - 1982
B.Sc. Chemistry, Leiden University, The Netherlands
Publications
(Recent publications)
Harnicek, D., Kampmann, E., Lauber, K., Hennel, R., Cardoso Martins, A.S., Guo, Y., Belka, C., Mörtl, S., Gallmeier, E., Kanaar, R., Mansmann, U., Lindner, L.H. and Issels, R.D. (2016). Hyperthermia adds to trabectedin effectiveness and thermal enhancement is associated with BRCA2 degradation and impairment of DNA homologous recombination repair. Int. J. Cancer, doi: 10.1002/ijc.30070.
Naipal, K.A., Verkaik, N.S., Sánchez, H., van Deurzen, C.H., den Bakker, M.A., Hoeijmakers, J.H., Kanaar, R., Vreeswijk, M.P., Jager, A. and van Gent, D.C. (2016). Tumor slice culture system to assess drug response of primary breast cancer. BMC Cancer 16, 78.
Issels, R., Kampmann, E., Kanaar, R. and Lindner, L.H. (2016). Hallmarks of hyperthermia in driving the future of clinical hyperthermia as targeted therapy: translation into clinical application. Int. J. Hyperthermia Jan 24, 1-7.
Tham, K.C., Kanaar, R. and Lebbink, J.H. (2015). Mismatch repair and homeologous recombination. DNA Repair 38, 75-83.
Ameziane, N., May, P., Haitjema, A., Van de Vrugt, H.J., Van Rossum-Fikkert, S.E., Ristic, D., Williams, G.J., Balk, J., Rockx, D., Li, H., Rooimans, M.A., Oostra, A.B., Velleuer, E., Dietrich, R., Bleijerveld, O.B., Altelaar, A.F.M., Meijers-Heijboer, H., Joenje, H., Glusman, G., Roach, J., Hood, L., Galas, D., Wyman, C., Balling, R., den Dunnen, J., De Winter, J.P., Kanaar, R., Gelinas, R. and Dorsman, J.C. (2015). A novel Fanconi anemia subtype associated with a dominant-negative mutation in RAD51. Nature Comm. 6, 8829.
Ramnath, N.W., Hawinkels, L.J., van Heijningen, P.M., te Riet, L., Paauwe, M., Kanaar, R., ten Dijke, P. and Essers, J. (2015). Fibulin-4 deficiency results in enhanced TGF-β signaling in aortic smooth muscle cells and increased TGF-β2 secretion. Sci. Rep. 5, 16872.
Naipal, K.A., Raams, A., Bruens, S.T., Verkaik, N.S., Jaspers, N.G., Hoeijmakers, J.H., van Leenders, A., Pothof, J., Kanaar, R., Boormans, J. and van Gent, D.C. (2015). Attenuated XPC expression does not associate with impaired DNA repair in bladder cancer. PLoS One 10, e0126029.
Braks, J.A., Spiegelberg. L., Koljenovic, S., Ridwan, Y., Keereweer, S., Kanaar, R., Wolvius, E.B. and Essers, J. (2015). Optical imaging of tumor response to hyperbaric oxygen treatment and irradiation in an orthotopic mouse model of head and neck squamous cell carcinoma. Mol. Imaging Biol. 7, 633-642.
Reuter, M., Zelensky, A., Smal, I., Meijering, E., van Cappellen, W.A., de Gruiter, M.H., van Belle, G.J., van Royen, M.E., Houtsmuller, A.B., Essers, J., Kanaar, R. and Wyman, C. (2014). BRCA2 diffuses as oligomeric clusters with RAD51 and changes mobility after DNA damage in live cells. J. Cell Biol. 207, 599-613.
Uringa, E.J., Baldeyron, C., Odijk, H., Wassenaar, E., van Cappellen, W.A., Maas, A., Hoeijmakers, J.H., Baarends, W.M., Kanaar, R. and Essers, J. (2015). A mRad51-GFP antimorphic allele affects homologous recombination and DNA damage sensitivity. DNA Repair 25, 27-40.
Berghauser Pont, L.M.E., Kloezeman, J.J., Naipal, K., van den Bent, M., van Gent, D.C., Dirven, C.M.F., Kanaar, R., Lamfers, M.L. and Leenstra, S. (2015). DNA damage response and anti-apoptotic proteins are predictors of histone deacetylase inhibitors-mediated radiosensitization of glioblastomas. Cancer Res. 28, 525-535.
Zelensky, A., Kanaar, R. and Wyman, C. (2014). Mediators of homologous DNA pairing. Cold Spring Harb. Perspect. Biol. 6, a016451.
Ramnath, N.W.M., van de Luijtgaarden, K.M., van der Pluijm, I., van Nimwegen, M., van Heijningen, P., Swagemakers, S., van Thiel, B., Ridwan, R., van Vliet N., Vermeij, M., Hawinkels, L., de Munck, A., Dzyubachyk, O., Meijering, E., van der Spek, P., Rottier, R., Hendriks, R.W., Kanaar, R., Rouwet, E.V., Kleinjan, A. and Essers, J. (2014). Extracellular matrix defects in aneurysmal Fibulin-4 deficient mice predispose to lung emphysema. PloS One 9, e106054.
Naipal, K.A., Verkaik, N.S., Ameziane, N., van Deurzen, C.H., Ter Brugge, P., Meijers, M., Sieuwerts, A.M., Martens, J., O'Connor, M.J., Vrieling, H., Hoeijmakers, J.H., Jonkers, J., Kanaar, R., de Winter, J., Vreeswijk, M., Jager, A. and van Gent, D.C. (2014). Functional ex vivo assay to select homologus recombination deficient breast tumors for PARP inhibitor treatment. Clin. Cancer Res. 20, 4816-4826.
Kuzniar, A. and Kanaar, R. (2014). PIQMIe: a web server for semi-quantitative proteomics data management and analysis. Nucleic Acids Res. 42, W100-106.
Carvalho, J.F. and Kanaar, R. (2014). Targeting homologous recombination-mediated DNA repair in cancer. Expert Opin. Ther. Targets 18, 427-458.
Shima, H., Suzuki, H., Sun, J., Kono, K., Shi, L., Kinomura, A., Horikoshi, Y., Ikura, T., Ikura, M., Kanaar, R., Igarashi, K., Saitoh, H., Kurumizaka, H. and Tashiro, S. (2014). Activation of the SUMO modification system is required for the accumulation of RAD51 at sites of DNA damage. J. Cell Sci. 126, 5284-5292.
Teaching activities
Opportunities for BEP and MEP projects
(1) Biochemistry of homologous recombination
(2) Cell biology of homologous recombination
(3) DNA damage response to ultra low doses of radiation
(4) Heat to augment cancer therapy
(5) The DNA damage response as a cancer therapy selection tool
(6) Cancer-on-chip
Biochemistry of homologous recombination
Relevant publications
Ameziane et al., A novel Fanconi anemia subtype associated with a dominant-negative mutation in RAD51. Nature Comm. 6, 8829.
Zelensky et al., Caffeine suppresses homologous recombination through interference with RAD51-mediated joint molecule formation. Nucleic Acids Res. 41, 6475-6489.
Sanchez et al., Combined optical and topographic imaging reveals different arrangements of human RAD54 with presynaptic and postsynaptic RAD51-DNA filaments. Proc. Natl. Acad. Sci. USA 110, 11385-11390.
Ristic et al., Visualizing RAD51-mediated joint molecules: implications for recombination mechanism and the effect of sequence heterology. Nucleic Acids Res. 39, 155-167.
Holthausen et al., Effect of the BRCA2 CTRD domain on RAD51 filaments analyzed by an ensemble of single molecule techniques. Nucleic Acids Res. 39, 6558-6567.
Cell biology of homologous recombination
Relevant publications:
Reuter and Zelensky et al., BRCA2 diffuses as oligomeric clusters with RAD51 and changes mobility after DNA damage in live cells. J. Cell Biol. 207, 599-613.
Uringa et al., A mRad51-GFP antimorphic allele affects homologous recombination and DNA damage sensitivity. DNA Repair 25, 27-40.
Agarwal et al., ATP-dependent and ATP-independent functions of Rad54 in genome maintenance. J. Cell Biol. 192, 735-750.
DNA damage response to ultra low doses of radiation
Insertional mutagenesis occurs when extrachromosomal DNA integrates in the host’s genome. Integration of exogenous DNA is an important factor in several therapeutic approaches, where it is regarded as either beneficial (stable restoration of a missing gene) or dangerous (insertion near an oncogene and its activation). Indeed, viral integration is considered a contributing factor in oncogenesis, even for viruses that do not encode an active integration function.
We discovered that ionizing radiation doses as low at 0.01 Gy significantly increase insertion of exogenous DNA in the genome of mammalian cells. Several of our observations challenge the intuitive explanation of the phenomenon as trapping an extrachromsomal DNA fragment during mis-repair of a genomic double-strand break by non-homologous end joining.
Importantly, we showed that that the extremely low doses of radiation also promoted the insertion of DNA from (non-integrating) viruses residing in the cells. Because the doses tested are in the range of those received by patients undergoing a computed tomography (CT) scan (10 mGy for an abdominal scan), we also performed experiments in a CT scanner and again observed the same increase in insertional mutagenesis. Given that an estimated 68 million CT scans per year are performed in the USA and the prevalence of persistent viral infections, it is of great importance to unravel the molecular mechanism of the extremely low dose induced insertional mutagenesis pathway.
Using state-of-the art genome engineering and cell biological experiments we are unraveling the molecular mechanism of this novel DNA damage response mechanism.
Relevant publications
In preparation
Heat to augment cancer therapy
We discovered that local mild hyperthermia attenuates the DNA damage response by inhibiting homologous recombination through degradation of the BRCA2 protein. Our discovery explains the clinical observation that hyperthermia sensitizes tumors to DNA damage-inducing therapies. We showed that mild hyperthermia sensitizes innately homologous-proficient tumors to PARP-1 inhibitors, thus irrespective of genetic BRCA deficiency. These finding are very promising as they suggest that use of PARP-1 inhibitor precision medicines can be extended beyond the limited group of patient who are BRCA mutation carriers. Here we want to find define the molecular mechanism through which the BRCA2 is degraded and make use of that knowledge to guide the design of optimize hyperthermia protocols. We will employ molecular biology, quantitative cell biology and proteomics techniques.
Relevant publications
Krawczyk et al., Mild hyperthermia inhibits homologous recombination, induces BRCA2 degradation, and sensitizes cancer cells to poly (ADP-ribose) polymerase-1 inhibition. Proc. Natl. Acad. Sci. USA 108, 9851-9856.
van den Tempel et al., Improving efficacy of hyperthermia in oncology by exploiting biological mechanisms. Int. J. Hyperthermia, in press.
The DNA damage response as a cancer therapy selection tool
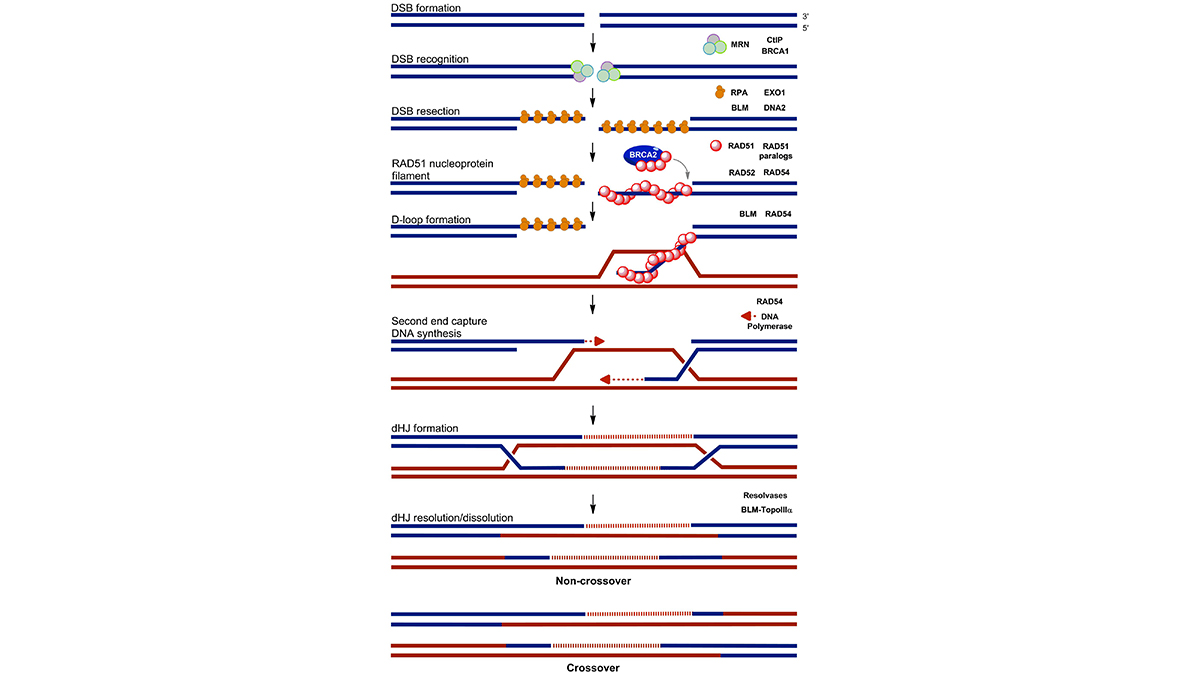
To treat cancer patients successfully and with minimal toxic side effects precision therapies are of utmost importance. Our research program address challenges that can be identified in taking personalized cancer treatments forward, specifically developing better patient selection methods. Rooted in our experience in analyzing mechanisms of the DNA damage response and in particular the homologous recombination DNA repair pathway, we expect to widen the applicability of the successful concept of the synthetic lethality due to PARP-1 inhibition in BRCA-deficient tumors. This research program will result in optimized functional ex vivo assays for homologous recombination deficiency on fresh breast cancer tumor material. We are working to adopt the assay to other tumor sites. Importantly, we aim to correlate the outcome of the ex vivo DNA damage response assays and tumor response to PARP-1 inhibitor and/or cisplatin treatment in breast cancer patients. Furthermore, our functional ex vivo DNA damage response assays can be used to uncover novel biomarkers for homologous recombination deficiency with the use of NGS methods and bioinformatics. As a result many more patients that would benefit from therapies that target homologous recombination deficiency can be selected. See also: The DNA damage response in tissues and tumors.
Relevant publications
Naipal et al., Functional ex vivo assay to select homologus recombination deficient breast tumors for PARP inhibitor treatment. Clin. Cancer Res. 20, 4816-4826.
Naipal et al., Tumor slice culture system to assess drug response of primary breast cancer. BMC Cancer 16, 78.
Cancer-on-chip
This is a biomedical engineering projec in collabaration between the Erasmus MC (Dik van Gent), TU Delft (Ronald Dekker) and Philips Research (Anja van der Stolpe).
We are developing the first generation cancer-on-chip models for functional precision diagnostic purposes: microfluidic chips containing cultures of viable slices from resected primary breast cancer tumors to test drug efficacy on an individual patient basis. The organ-on-chip approach allows improved mimicking of the ex vivo cancer cell microenvironment, both with respect to physical and biochemical factors, allowing optimal primary cancer cell culture under tightly controlled growth conditions. We aim to provide the first proof of principle that primary cancer culture in an organ-on-chip device can have clinical utility as a diagnostic test to functionally assess the response of an individual patient’s tumor to a targeted drug.
Relevant publications:
Work in progress






